4.10. Predicting House Prices on Kaggle¶ Open the notebook in SageMaker Studio Lab
Now that we have introduced some basic tools for building and training deep networks and regularizing them with techniques including weight decay and dropout, we are ready to put all this knowledge into practice by participating in a Kaggle competition. The house price prediction competition is a great place to start. The data are fairly generic and do not exhibit exotic structure that might require specialized models (as audio or video might). This dataset, collected by Bart de Cock in 2011 (), covers house prices in Ames, IA from the period of 2006–2010. It is considerably larger than the famous Boston housing dataset of Harrison and Rubinfeld (1978), boasting both more examples and more features.
In this section, we will walk you through details of data preprocessing, model design, and hyperparameter selection. We hope that through a hands-on approach, you will gain some intuitions that will guide you in your career as a data scientist.
4.10.1. Downloading and Caching Datasets¶
Throughout the book, we will train and test models on various downloaded
datasets. Here, we implement several utility functions to facilitate
data downloading. First, we maintain a dictionary DATA_HUB that maps
a string (the name of the dataset) to a tuple containing both the URL
to locate the dataset and the SHA-1 key that verifies the integrity of
the file. All such datasets are hosted at the site whose address is
DATA_URL.
import hashlib
import os
import tarfile
import zipfile
import requests
#@save
DATA_HUB = dict()
DATA_URL = 'http://d2l-data.s3-accelerate.amazonaws.com/'
The following download function downloads a dataset, caches it in a
local directory (../data by default), and returns the name of the
downloaded file. If a file corresponding to this dataset already exists
in the cache directory and its SHA-1 matches the one stored in
DATA_HUB, our code will use the cached file to avoid clogging up
your internet with redundant downloads.
def download(name, cache_dir=os.path.join('..', 'data')): #@save
"""Download a file inserted into DATA_HUB, return the local filename."""
assert name in DATA_HUB, f"{name} does not exist in {DATA_HUB}."
url, sha1_hash = DATA_HUB[name]
os.makedirs(cache_dir, exist_ok=True)
fname = os.path.join(cache_dir, url.split('/')[-1])
if os.path.exists(fname):
sha1 = hashlib.sha1()
with open(fname, 'rb') as f:
while True:
data = f.read(1048576)
if not data:
break
sha1.update(data)
if sha1.hexdigest() == sha1_hash:
return fname # Hit cache
print(f'Downloading {fname} from {url}...')
r = requests.get(url, stream=True, verify=True)
with open(fname, 'wb') as f:
f.write(r.content)
return fname
We also implement two additional utility functions: one is to download
and extract a zip or tar file and the other to download all the datasets
used in this book from DATA_HUB into the cache directory.
def download_extract(name, folder=None): #@save
"""Download and extract a zip/tar file."""
fname = download(name)
base_dir = os.path.dirname(fname)
data_dir, ext = os.path.splitext(fname)
if ext == '.zip':
fp = zipfile.ZipFile(fname, 'r')
elif ext in ('.tar', '.gz'):
fp = tarfile.open(fname, 'r')
else:
assert False, 'Only zip/tar files can be extracted.'
fp.extractall(base_dir)
return os.path.join(base_dir, folder) if folder else data_dir
def download_all(): #@save
"""Download all files in the DATA_HUB."""
for name in DATA_HUB:
download(name)
4.10.2. Kaggle¶
Kaggle is a popular platform that hosts machine learning competitions. Each competition centers on a dataset and many are sponsored by stakeholders who offer prizes to the winning solutions. The platform helps users to interact via forums and shared code, fostering both collaboration and competition. While leaderboard chasing often spirals out of control, with researchers focusing myopically on preprocessing steps rather than asking fundamental questions, there is also tremendous value in the objectivity of a platform that facilitates direct quantitative comparisons among competing approaches as well as code sharing so that everyone can learn what did and did not work. If you want to participate in a Kaggle competition, you will first need to register for an account (see Fig. 4.10.1).
Fig. 4.10.1 The Kaggle website.¶
On the house price prediction competition page, as illustrated in Fig. 4.10.2, you can find the dataset (under the “Data” tab), submit predictions, and see your ranking, The URL is right here:
Fig. 4.10.2 The house price prediction competition page.¶
4.10.3. Accessing and Reading the Dataset¶
Note that the competition data is separated into training and test sets. Each record includes the property value of the house and attributes such as street type, year of construction, roof type, basement condition, etc. The features consist of various data types. For example, the year of construction is represented by an integer, the roof type by discrete categorical assignments, and other features by floating point numbers. And here is where reality complicates things: for some examples, some data are altogether missing with the missing value marked simply as “na”. The price of each house is included for the training set only (it is a competition after all). We will want to partition the training set to create a validation set, but we only get to evaluate our models on the official test set after uploading predictions to Kaggle. The “Data” tab on the competition tab in Fig. 4.10.2 has links to download the data.
To get started, we will read in and process the data using pandas,
which we have introduced in Section 2.2. So, you will want to
make sure that you have pandas installed before proceeding further.
Fortunately, if you are reading in Jupyter, we can install pandas
without even leaving the notebook.
# If pandas is not installed, please uncomment the following line:
# !pip install pandas
%matplotlib inline
import pandas as pd
from mxnet import autograd, gluon, init, np, npx
from mxnet.gluon import nn
from d2l import mxnet as d2l
npx.set_np()
# If pandas is not installed, please uncomment the following line:
# !pip install pandas
%matplotlib inline
import numpy as np
import pandas as pd
import torch
from torch import nn
from d2l import torch as d2l
# If pandas is not installed, please uncomment the following line:
# !pip install pandas
%matplotlib inline
import numpy as np
import pandas as pd
import tensorflow as tf
from d2l import tensorflow as d2l
For convenience, we can download and cache the Kaggle housing dataset using the script we defined above.
DATA_HUB['kaggle_house_train'] = ( #@save
DATA_URL + 'kaggle_house_pred_train.csv',
'585e9cc93e70b39160e7921475f9bcd7d31219ce')
DATA_HUB['kaggle_house_test'] = ( #@save
DATA_URL + 'kaggle_house_pred_test.csv',
'fa19780a7b011d9b009e8bff8e99922a8ee2eb90')
We use pandas to load the two csv files containing training and test
data respectively.
train_data = pd.read_csv(download('kaggle_house_train'))
test_data = pd.read_csv(download('kaggle_house_test'))
Downloading ../data/kaggle_house_pred_train.csv from http://d2l-data.s3-accelerate.amazonaws.com/kaggle_house_pred_train.csv...
Downloading ../data/kaggle_house_pred_test.csv from http://d2l-data.s3-accelerate.amazonaws.com/kaggle_house_pred_test.csv...
train_data = pd.read_csv(download('kaggle_house_train'))
test_data = pd.read_csv(download('kaggle_house_test'))
Downloading ../data/kaggle_house_pred_train.csv from http://d2l-data.s3-accelerate.amazonaws.com/kaggle_house_pred_train.csv...
Downloading ../data/kaggle_house_pred_test.csv from http://d2l-data.s3-accelerate.amazonaws.com/kaggle_house_pred_test.csv...
train_data = pd.read_csv(download('kaggle_house_train'))
test_data = pd.read_csv(download('kaggle_house_test'))
Downloading ../data/kaggle_house_pred_train.csv from http://d2l-data.s3-accelerate.amazonaws.com/kaggle_house_pred_train.csv...
Downloading ../data/kaggle_house_pred_test.csv from http://d2l-data.s3-accelerate.amazonaws.com/kaggle_house_pred_test.csv...
The training dataset includes 1460 examples, 80 features, and 1 label, while the test data contains 1459 examples and 80 features.
print(train_data.shape)
print(test_data.shape)
(1460, 81)
(1459, 80)
print(train_data.shape)
print(test_data.shape)
(1460, 81)
(1459, 80)
print(train_data.shape)
print(test_data.shape)
(1460, 81)
(1459, 80)
Let us take a look at the first four and last two features as well as the label (SalePrice) from the first four examples.
print(train_data.iloc[0:4, [0, 1, 2, 3, -3, -2, -1]])
Id MSSubClass MSZoning LotFrontage SaleType SaleCondition SalePrice
0 1 60 RL 65.0 WD Normal 208500
1 2 20 RL 80.0 WD Normal 181500
2 3 60 RL 68.0 WD Normal 223500
3 4 70 RL 60.0 WD Abnorml 140000
print(train_data.iloc[0:4, [0, 1, 2, 3, -3, -2, -1]])
Id MSSubClass MSZoning LotFrontage SaleType SaleCondition SalePrice
0 1 60 RL 65.0 WD Normal 208500
1 2 20 RL 80.0 WD Normal 181500
2 3 60 RL 68.0 WD Normal 223500
3 4 70 RL 60.0 WD Abnorml 140000
print(train_data.iloc[0:4, [0, 1, 2, 3, -3, -2, -1]])
Id MSSubClass MSZoning LotFrontage SaleType SaleCondition SalePrice
0 1 60 RL 65.0 WD Normal 208500
1 2 20 RL 80.0 WD Normal 181500
2 3 60 RL 68.0 WD Normal 223500
3 4 70 RL 60.0 WD Abnorml 140000
We can see that in each example, the first feature is the ID. This helps the model identify each training example. While this is convenient, it does not carry any information for prediction purposes. Hence, we remove it from the dataset before feeding the data into the model.
all_features = pd.concat((train_data.iloc[:, 1:-1], test_data.iloc[:, 1:]))
4.10.4. Data Preprocessing¶
As stated above, we have a wide variety of data types. We will need to preprocess the data before we can start modeling. Let us start with the numerical features. First, we apply a heuristic, replacing all missing values by the corresponding feature’s mean. Then, to put all features on a common scale, we standardize the data by rescaling features to zero mean and unit variance:
where \(\mu\) and \(\sigma\) denote mean and standard deviation, respectively. To verify that this indeed transforms our feature (variable) such that it has zero mean and unit variance, note that \(E[\frac{x-\mu}{\sigma}] = \frac{\mu - \mu}{\sigma} = 0\) and that \(E[(x-\mu)^2] = (\sigma^2 + \mu^2) - 2\mu^2+\mu^2 = \sigma^2\). Intuitively, we standardize the data for two reasons. First, it proves convenient for optimization. Second, because we do not know a priori which features will be relevant, we do not want to penalize coefficients assigned to one feature more than on any other.
# If test data were inaccessible, mean and standard deviation could be
# calculated from training data
numeric_features = all_features.dtypes[all_features.dtypes != 'object'].index
all_features[numeric_features] = all_features[numeric_features].apply(
lambda x: (x - x.mean()) / (x.std()))
# After standardizing the data all means vanish, hence we can set missing
# values to 0
all_features[numeric_features] = all_features[numeric_features].fillna(0)
Next we deal with discrete values. This includes features such as
“MSZoning”. We replace them by a one-hot encoding in the same way that
we previously transformed multiclass labels into vectors (see
Section 3.4.1). For instance, “MSZoning”
assumes the values “RL” and “RM”. Dropping the “MSZoning” feature, two
new indicator features “MSZoning_RL” and “MSZoning_RM” are created
with values being either 0 or 1. According to one-hot encoding, if the
original value of “MSZoning” is “RL”, then “MSZoning_RL” is 1 and
“MSZoning_RM” is 0. The pandas package does this automatically for
us.
# `Dummy_na=True` considers "na" (missing value) as a valid feature value, and
# creates an indicator feature for it
all_features = pd.get_dummies(all_features, dummy_na=True)
all_features.shape
(2919, 331)
# `Dummy_na=True` considers "na" (missing value) as a valid feature value, and
# creates an indicator feature for it
all_features = pd.get_dummies(all_features, dummy_na=True)
all_features.shape
(2919, 331)
# `Dummy_na=True` considers "na" (missing value) as a valid feature value, and
# creates an indicator feature for it
all_features = pd.get_dummies(all_features, dummy_na=True)
all_features.shape
(2919, 331)
You can see that this conversion increases the number of features from
79 to 331. Finally, via the values attribute, we can extract the
NumPy format from the pandas format and convert it into the tensor
representation for training.
n_train = train_data.shape[0]
train_features = np.array(all_features[:n_train].values, dtype=np.float32)
test_features = np.array(all_features[n_train:].values, dtype=np.float32)
train_labels = np.array(
train_data.SalePrice.values.reshape(-1, 1), dtype=np.float32)
n_train = train_data.shape[0]
train_features = torch.tensor(all_features[:n_train].values, dtype=torch.float32)
test_features = torch.tensor(all_features[n_train:].values, dtype=torch.float32)
train_labels = torch.tensor(
train_data.SalePrice.values.reshape(-1, 1), dtype=torch.float32)
n_train = train_data.shape[0]
train_features = tf.constant(all_features[:n_train].values, dtype=tf.float32)
test_features = tf.constant(all_features[n_train:].values, dtype=tf.float32)
train_labels = tf.constant(
train_data.SalePrice.values.reshape(-1, 1), dtype=tf.float32)
4.10.5. Training¶
To get started we train a linear model with squared loss. Not surprisingly, our linear model will not lead to a competition-winning submission but it provides a sanity check to see whether there is meaningful information in the data. If we cannot do better than random guessing here, then there might be a good chance that we have a data processing bug. And if things work, the linear model will serve as a baseline giving us some intuition about how close the simple model gets to the best reported models, giving us a sense of how much gain we should expect from fancier models.
loss = gluon.loss.L2Loss()
def get_net():
net = nn.Sequential()
net.add(nn.Dense(1))
net.initialize()
return net
loss = nn.MSELoss()
in_features = train_features.shape[1]
def get_net():
net = nn.Sequential(nn.Linear(in_features,1))
return net
loss = tf.keras.losses.MeanSquaredError()
def get_net():
net = tf.keras.models.Sequential()
net.add(tf.keras.layers.Dense(
1, kernel_regularizer=tf.keras.regularizers.l2(weight_decay)))
return net
With house prices, as with stock prices, we care about relative quantities more than absolute quantities. Thus we tend to care more about the relative error \(\frac{y - \hat{y}}{y}\) than about the absolute error \(y - \hat{y}\). For instance, if our prediction is off by USD 100,000 when estimating the price of a house in Rural Ohio, where the value of a typical house is 125,000 USD, then we are probably doing a horrible job. On the other hand, if we err by this amount in Los Altos Hills, California, this might represent a stunningly accurate prediction (there, the median house price exceeds 4 million USD).
One way to address this problem is to measure the discrepancy in the logarithm of the price estimates. In fact, this is also the official error measure used by the competition to evaluate the quality of submissions. After all, a small value \(\delta\) for \(|\log y - \log \hat{y}| \leq \delta\) translates into \(e^{-\delta} \leq \frac{\hat{y}}{y} \leq e^\delta\). This leads to the following root-mean-squared-error between the logarithm of the predicted price and the logarithm of the label price:
def log_rmse(net, features, labels):
# To further stabilize the value when the logarithm is taken, set the
# value less than 1 as 1
clipped_preds = np.clip(net(features), 1, float('inf'))
return np.sqrt(2 * loss(np.log(clipped_preds), np.log(labels)).mean())
def log_rmse(net, features, labels):
# To further stabilize the value when the logarithm is taken, set the
# value less than 1 as 1
clipped_preds = torch.clamp(net(features), 1, float('inf'))
rmse = torch.sqrt(loss(torch.log(clipped_preds),
torch.log(labels)))
return rmse.item()
def log_rmse(y_true, y_pred):
# To further stabilize the value when the logarithm is taken, set the
# value less than 1 as 1
clipped_preds = tf.clip_by_value(y_pred, 1, float('inf'))
return tf.sqrt(tf.reduce_mean(loss(
tf.math.log(y_true), tf.math.log(clipped_preds))))
Unlike in previous sections, our training functions will rely on the Adam optimizer (we will describe it in greater detail later). The main appeal of this optimizer is that, despite doing no better (and sometimes worse) given unlimited resources for hyperparameter optimization, people tend to find that it is significantly less sensitive to the initial learning rate.
def train(net, train_features, train_labels, test_features, test_labels,
num_epochs, learning_rate, weight_decay, batch_size):
train_ls, test_ls = [], []
train_iter = d2l.load_array((train_features, train_labels), batch_size)
# The Adam optimization algorithm is used here
trainer = gluon.Trainer(net.collect_params(), 'adam', {
'learning_rate': learning_rate, 'wd': weight_decay})
for epoch in range(num_epochs):
for X, y in train_iter:
with autograd.record():
l = loss(net(X), y)
l.backward()
trainer.step(batch_size)
train_ls.append(log_rmse(net, train_features, train_labels))
if test_labels is not None:
test_ls.append(log_rmse(net, test_features, test_labels))
return train_ls, test_ls
def train(net, train_features, train_labels, test_features, test_labels,
num_epochs, learning_rate, weight_decay, batch_size):
train_ls, test_ls = [], []
train_iter = d2l.load_array((train_features, train_labels), batch_size)
# The Adam optimization algorithm is used here
optimizer = torch.optim.Adam(net.parameters(),
lr = learning_rate,
weight_decay = weight_decay)
for epoch in range(num_epochs):
for X, y in train_iter:
optimizer.zero_grad()
l = loss(net(X), y)
l.backward()
optimizer.step()
train_ls.append(log_rmse(net, train_features, train_labels))
if test_labels is not None:
test_ls.append(log_rmse(net, test_features, test_labels))
return train_ls, test_ls
def train(net, train_features, train_labels, test_features, test_labels,
num_epochs, learning_rate, weight_decay, batch_size):
train_ls, test_ls = [], []
train_iter = d2l.load_array((train_features, train_labels), batch_size)
# The Adam optimization algorithm is used here
optimizer = tf.keras.optimizers.Adam(learning_rate)
net.compile(loss=loss, optimizer=optimizer)
for epoch in range(num_epochs):
for X, y in train_iter:
with tf.GradientTape() as tape:
y_hat = net(X)
l = loss(y, y_hat)
params = net.trainable_variables
grads = tape.gradient(l, params)
optimizer.apply_gradients(zip(grads, params))
train_ls.append(log_rmse(train_labels, net(train_features)))
if test_labels is not None:
test_ls.append(log_rmse(test_labels, net(test_features)))
return train_ls, test_ls
4.10.6. \(K\)-Fold Cross-Validation¶
You might recall that we introduced \(K\)-fold cross-validation in the section where we discussed how to deal with model selection (Section 4.4). We will put this to good use to select the model design and to adjust the hyperparameters. We first need a function that returns the \(i^\mathrm{th}\) fold of the data in a \(K\)-fold cross-validation procedure. It proceeds by slicing out the \(i^\mathrm{th}\) segment as validation data and returning the rest as training data. Note that this is not the most efficient way of handling data and we would definitely do something much smarter if our dataset was considerably larger. But this added complexity might obfuscate our code unnecessarily so we can safely omit it here owing to the simplicity of our problem.
def get_k_fold_data(k, i, X, y):
assert k > 1
fold_size = X.shape[0] // k
X_train, y_train = None, None
for j in range(k):
idx = slice(j * fold_size, (j + 1) * fold_size)
X_part, y_part = X[idx, :], y[idx]
if j == i:
X_valid, y_valid = X_part, y_part
elif X_train is None:
X_train, y_train = X_part, y_part
else:
X_train = np.concatenate([X_train, X_part], 0)
y_train = np.concatenate([y_train, y_part], 0)
return X_train, y_train, X_valid, y_valid
def get_k_fold_data(k, i, X, y):
assert k > 1
fold_size = X.shape[0] // k
X_train, y_train = None, None
for j in range(k):
idx = slice(j * fold_size, (j + 1) * fold_size)
X_part, y_part = X[idx, :], y[idx]
if j == i:
X_valid, y_valid = X_part, y_part
elif X_train is None:
X_train, y_train = X_part, y_part
else:
X_train = torch.cat([X_train, X_part], 0)
y_train = torch.cat([y_train, y_part], 0)
return X_train, y_train, X_valid, y_valid
def get_k_fold_data(k, i, X, y):
assert k > 1
fold_size = X.shape[0] // k
X_train, y_train = None, None
for j in range(k):
idx = slice(j * fold_size, (j + 1) * fold_size)
X_part, y_part = X[idx, :], y[idx]
if j == i:
X_valid, y_valid = X_part, y_part
elif X_train is None:
X_train, y_train = X_part, y_part
else:
X_train = tf.concat([X_train, X_part], 0)
y_train = tf.concat([y_train, y_part], 0)
return X_train, y_train, X_valid, y_valid
The training and verification error averages are returned when we train \(K\) times in the \(K\)-fold cross-validation.
def k_fold(k, X_train, y_train, num_epochs, learning_rate, weight_decay,
batch_size):
train_l_sum, valid_l_sum = 0, 0
for i in range(k):
data = get_k_fold_data(k, i, X_train, y_train)
net = get_net()
train_ls, valid_ls = train(net, *data, num_epochs, learning_rate,
weight_decay, batch_size)
train_l_sum += train_ls[-1]
valid_l_sum += valid_ls[-1]
if i == 0:
d2l.plot(list(range(1, num_epochs + 1)), [train_ls, valid_ls],
xlabel='epoch', ylabel='rmse', xlim=[1, num_epochs],
legend=['train', 'valid'], yscale='log')
print(f'fold {i + 1}, train log rmse {float(train_ls[-1]):f}, '
f'valid log rmse {float(valid_ls[-1]):f}')
return train_l_sum / k, valid_l_sum / k
4.10.7. Model Selection¶
In this example, we pick an untuned set of hyperparameters and leave it up to the reader to improve the model. Finding a good choice can take time, depending on how many variables one optimizes over. With a large enough dataset, and the normal sorts of hyperparameters, \(K\)-fold cross-validation tends to be reasonably resilient against multiple testing. However, if we try an unreasonably large number of options we might just get lucky and find that our validation performance is no longer representative of the true error.
k, num_epochs, lr, weight_decay, batch_size = 5, 100, 5, 0, 64
train_l, valid_l = k_fold(k, train_features, train_labels, num_epochs, lr,
weight_decay, batch_size)
print(f'{k}-fold validation: avg train log rmse: {float(train_l):f}, '
f'avg valid log rmse: {float(valid_l):f}')
[19:54:10] src/base.cc:49: GPU context requested, but no GPUs found.
fold 1, train log rmse 0.169786, valid log rmse 0.157285
fold 2, train log rmse 0.162134, valid log rmse 0.189370
fold 3, train log rmse 0.163498, valid log rmse 0.167806
fold 4, train log rmse 0.167417, valid log rmse 0.154570
fold 5, train log rmse 0.162476, valid log rmse 0.182716
5-fold validation: avg train log rmse: 0.165062, avg valid log rmse: 0.170349
k, num_epochs, lr, weight_decay, batch_size = 5, 100, 5, 0, 64
train_l, valid_l = k_fold(k, train_features, train_labels, num_epochs, lr,
weight_decay, batch_size)
print(f'{k}-fold validation: avg train log rmse: {float(train_l):f}, '
f'avg valid log rmse: {float(valid_l):f}')
fold 1, train log rmse 0.170585, valid log rmse 0.156633
fold 2, train log rmse 0.162439, valid log rmse 0.190557
fold 3, train log rmse 0.164291, valid log rmse 0.168361
fold 4, train log rmse 0.167880, valid log rmse 0.154581
fold 5, train log rmse 0.163331, valid log rmse 0.182985
5-fold validation: avg train log rmse: 0.165705, avg valid log rmse: 0.170624
k, num_epochs, lr, weight_decay, batch_size = 5, 100, 5, 0, 64
train_l, valid_l = k_fold(k, train_features, train_labels, num_epochs, lr,
weight_decay, batch_size)
print(f'{k}-fold validation: avg train log rmse: {float(train_l):f}, '
f'avg valid log rmse: {float(valid_l):f}')
fold 1, train log rmse 0.170455, valid log rmse 0.157146
fold 2, train log rmse 0.162041, valid log rmse 0.188886
fold 3, train log rmse 0.163909, valid log rmse 0.168482
fold 4, train log rmse 0.168148, valid log rmse 0.154647
fold 5, train log rmse 0.163174, valid log rmse 0.182978
5-fold validation: avg train log rmse: 0.165545, avg valid log rmse: 0.170428
Notice that sometimes the number of training errors for a set of hyperparameters can be very low, even as the number of errors on \(K\)-fold cross-validation is considerably higher. This indicates that we are overfitting. Throughout training you will want to monitor both numbers. Less overfitting might indicate that our data can support a more powerful model. Massive overfitting might suggest that we can gain by incorporating regularization techniques.
4.10.8. Submitting Predictions on Kaggle¶
Now that we know what a good choice of hyperparameters should be, we might as well use all the data to train on it (rather than just \(1-1/K\) of the data that are used in the cross-validation slices). The model that we obtain in this way can then be applied to the test set. Saving the predictions in a csv file will simplify uploading the results to Kaggle.
def train_and_pred(train_features, test_features, train_labels, test_data,
num_epochs, lr, weight_decay, batch_size):
net = get_net()
train_ls, _ = train(net, train_features, train_labels, None, None,
num_epochs, lr, weight_decay, batch_size)
d2l.plot(np.arange(1, num_epochs + 1), [train_ls], xlabel='epoch',
ylabel='log rmse', xlim=[1, num_epochs], yscale='log')
print(f'train log rmse {float(train_ls[-1]):f}')
# Apply the network to the test set
preds = net(test_features).asnumpy()
# Reformat it to export to Kaggle
test_data['SalePrice'] = pd.Series(preds.reshape(1, -1)[0])
submission = pd.concat([test_data['Id'], test_data['SalePrice']], axis=1)
submission.to_csv('submission.csv', index=False)
def train_and_pred(train_features, test_features, train_labels, test_data,
num_epochs, lr, weight_decay, batch_size):
net = get_net()
train_ls, _ = train(net, train_features, train_labels, None, None,
num_epochs, lr, weight_decay, batch_size)
d2l.plot(np.arange(1, num_epochs + 1), [train_ls], xlabel='epoch',
ylabel='log rmse', xlim=[1, num_epochs], yscale='log')
print(f'train log rmse {float(train_ls[-1]):f}')
# Apply the network to the test set
preds = net(test_features).detach().numpy()
# Reformat it to export to Kaggle
test_data['SalePrice'] = pd.Series(preds.reshape(1, -1)[0])
submission = pd.concat([test_data['Id'], test_data['SalePrice']], axis=1)
submission.to_csv('submission.csv', index=False)
def train_and_pred(train_features, test_features, train_labels, test_data,
num_epochs, lr, weight_decay, batch_size):
net = get_net()
train_ls, _ = train(net, train_features, train_labels, None, None,
num_epochs, lr, weight_decay, batch_size)
d2l.plot(np.arange(1, num_epochs + 1), [train_ls], xlabel='epoch',
ylabel='log rmse', xlim=[1, num_epochs], yscale='log')
print(f'train log rmse {float(train_ls[-1]):f}')
# Apply the network to the test set
preds = net(test_features).numpy()
# Reformat it to export to Kaggle
test_data['SalePrice'] = pd.Series(preds.reshape(1, -1)[0])
submission = pd.concat([test_data['Id'], test_data['SalePrice']], axis=1)
submission.to_csv('submission.csv', index=False)
One nice sanity check is to see whether the predictions on the test set
resemble those of the \(K\)-fold cross-validation process. If they
do, it is time to upload them to Kaggle. The following code will
generate a file called submission.csv.
train_and_pred(train_features, test_features, train_labels, test_data,
num_epochs, lr, weight_decay, batch_size)
train log rmse 0.162403
train_and_pred(train_features, test_features, train_labels, test_data,
num_epochs, lr, weight_decay, batch_size)
train log rmse 0.162490
train_and_pred(train_features, test_features, train_labels, test_data,
num_epochs, lr, weight_decay, batch_size)
train log rmse 0.162669
Next, as demonstrated in Fig. 4.10.3, we can submit our predictions on Kaggle and see how they compare with the actual house prices (labels) on the test set. The steps are quite simple:
Log in to the Kaggle website and visit the house price prediction competition page.
Click the “Submit Predictions” or “Late Submission” button (as of this writing, the button is located on the right).
Click the “Upload Submission File” button in the dashed box at the bottom of the page and select the prediction file you wish to upload.
Click the “Make Submission” button at the bottom of the page to view your results.
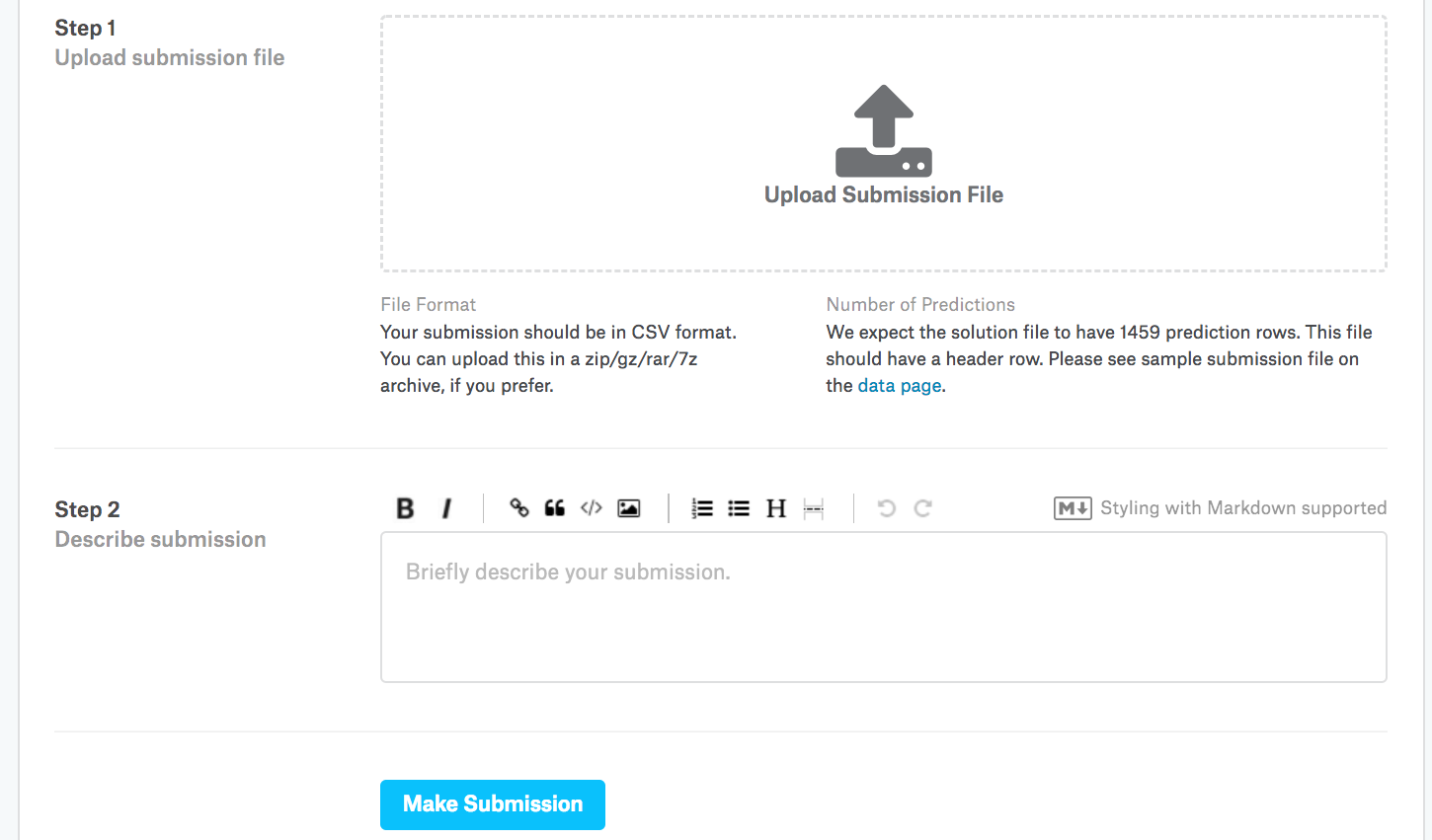
Fig. 4.10.3 Submitting data to Kaggle¶
4.10.9. Summary¶
Real data often contain a mix of different data types and need to be preprocessed.
Rescaling real-valued data to zero mean and unit variance is a good default. So is replacing missing values with their mean.
Transforming categorical features into indicator features allows us to treat them like one-hot vectors.
We can use \(K\)-fold cross-validation to select the model and adjust the hyperparameters.
Logarithms are useful for relative errors.
4.10.10. Exercises¶
Submit your predictions for this section to Kaggle. How good are your predictions?
Can you improve your model by minimizing the logarithm of prices directly? What happens if you try to predict the logarithm of the price rather than the price?
Is it always a good idea to replace missing values by their mean? Hint: can you construct a situation where the values are not missing at random?
Improve the score on Kaggle by tuning the hyperparameters through \(K\)-fold cross-validation.
Improve the score by improving the model (e.g., layers, weight decay, and dropout).
What happens if we do not standardize the continuous numerical features like what we have done in this section?
